Genetic mosaicism painting
Genetic mosaicism painting
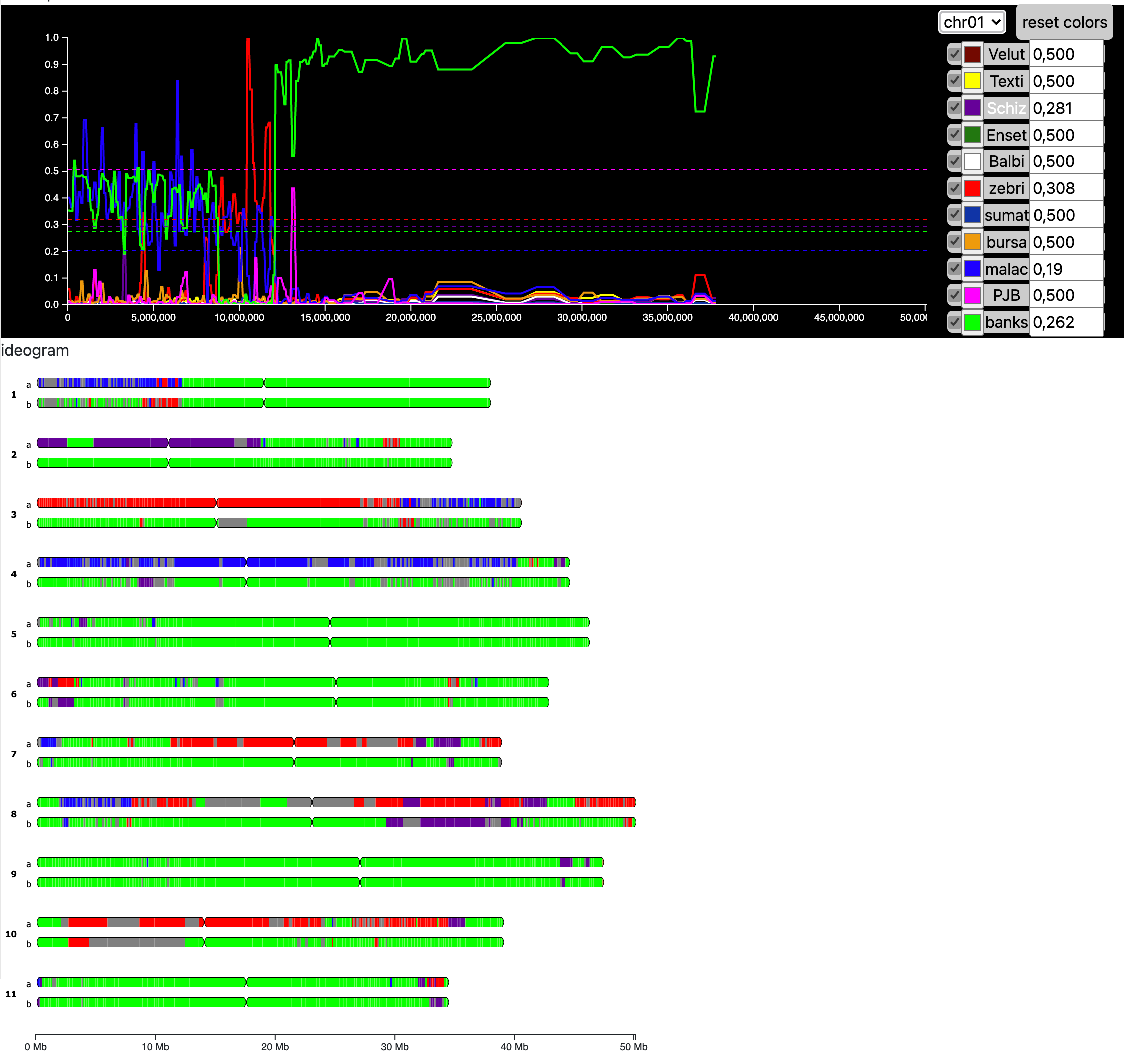
Mosaic visualization shows colored blocs on the chromosomes:Data curation displays the ancestral components along the chromosomes on an interactive graph to help the mosaïc construction:
Preloaded data from published studies or generated with VCFHunter or TraceAncestor.
Generate a visualization with your custom chromosomes data.
Chromosome data format, each column tab separated chr, len, centromereInf (optional), centromereSup (optional), label (optional) example : chr len centromereInf centromereSup label chr01 37945898 AB chr02 34728925 11000000 AB chr03 40528553 15000000 AB chr04 44598304 13000000 22000000 AB chr05 46248384 23000000 26000000 AB chr06 42818424 23000000 27000000 AB chr07 38870123 20000000 23000000 AB chr08 50124231 21000000 25000000 AB chr09 47435180 25000000 29000000 AB chr10 39038070 12000000 16000000 AB chr11 34441343 15000000 20000000 AB
Enter a block file to display the mosaïc visualization : chr haplotype start end ancestral_group 01 0 1 29070452 g4 01 1 1 29070452 g4 02 0 1 29511734 g4 02 1 1 29511734 g4Enter a file containing the ancestral components along the chromosomes to display an interactive graph and generate a new mosaïc visualisation : chr start end V T S chr01 1145 189582 0.001671983513048138 0.014082301923852172 0.0016386866759508464 chr01 189593 356965 0.0012441961973068657 0.012867234592909085 0.0018101312755326665 chr01 356968 488069 0.0011179557674005532 0.010035172902205201 0.000759432489203596 chr01 488097 633373 0.002678217164025965 0.010470727908771585 0.003896031529700906
Generating datasets In order to generate ready-to-use datasets, analyses can be conducted with the following software: * VCFHunter VCFHunter is a suite of python scripts enabling chromosome painting of indivisual based on the contribution of ancestral groups using VCF files. Please look at the tutorial: https://github.com/SouthGreenPlatform/VcfHunter/blob/master/turorial_painting_GEMO_visualization.md * TraceAncestor TraceAncestor permits to estimate the allelic dosage of ancestral alleles in hybrid individuals and then to perform chromosom painting. Please look at the tutorial: https://github.com/SouthGreenPlatform/TraceAncestor_gemo
group name hex un undef_group #b4b4b4 g6 group6 #c00000 g5 group5 #9a029a g3 group3 #1440cd g2 group2 #ffc000 g1 group1 #000000 g4 group4 #00b009 g7 group7 #b4b4b4 g0 group0 #000000
bed file example : chr01 5287838 5289224 gene 0 - chr01 15485703 15486813 gene 0 + chr01 15490016 15492587 gene 0 + chr02 2276353 2277821 gene 0 + chr02 15358393 15361764 gene 0 +
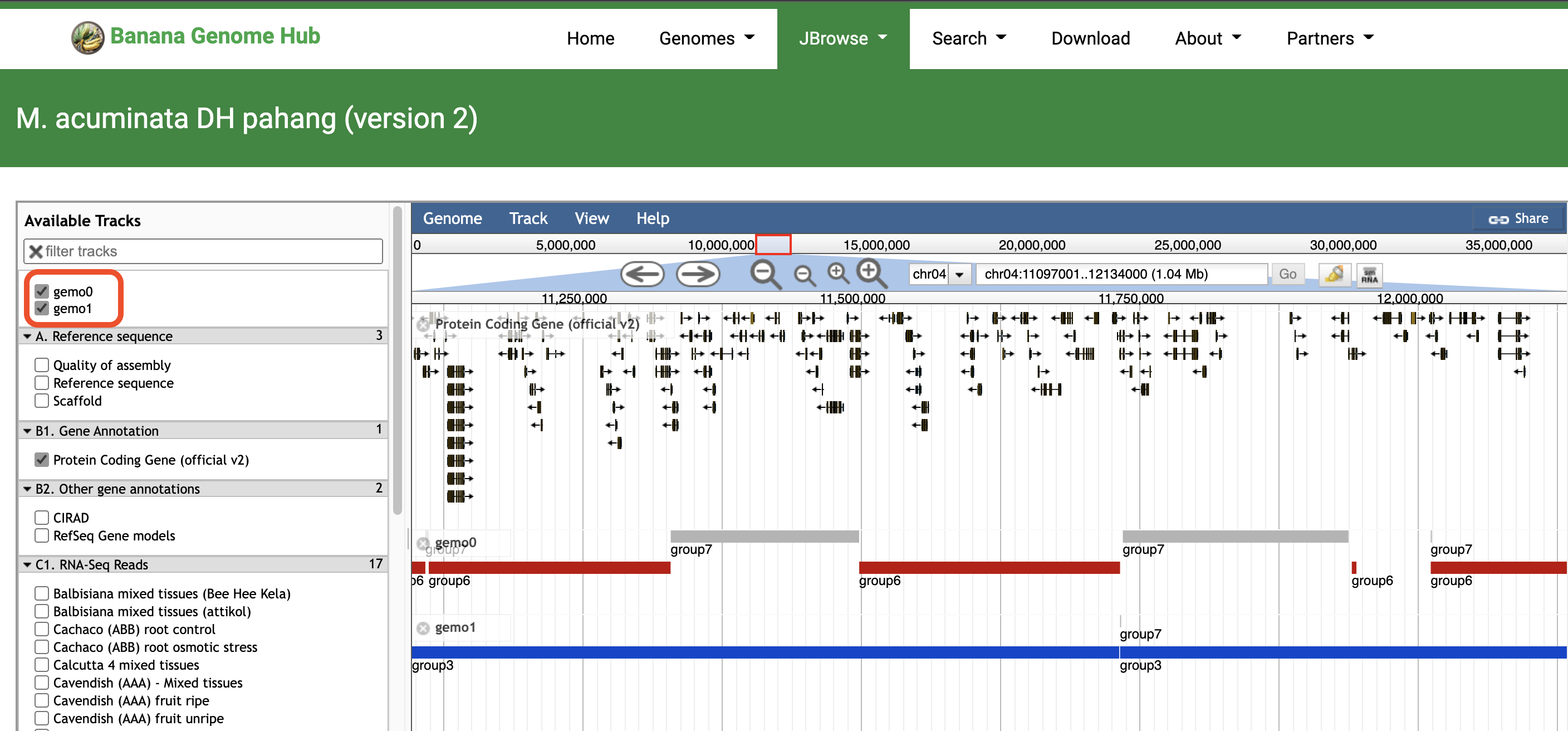
Jbrowse link example : https://banana-genome-hub.southgreen.fr/content/m-acuminata-dh-pahang-version-2/ GeMo will generate tooltips when hovering over each chromosomal block. A link is displayed to explore a chromosome block directly on jbrowse.The mosaic data are sent as tracks to be viewed in jbrowse.